DockIT
DockIT is a tool for interactive molecular docking
DockIT is software for interactive molecular docking. It enables the user to control the position and orientation of a ligand and a receptor bringing them into a docking pose either with a mouse and keyboard, with a haptic device or a VR headset (Oculus Rift, Rift S or Meta Quest2). Using a VR headset is the most immersive approach. Atomic interactions are modelled using molecular dynamics-based force-fields. It can be used for rigid docking and it also can be used incorporating receptor flexibility.
The following images are screenshots from the software.
 | 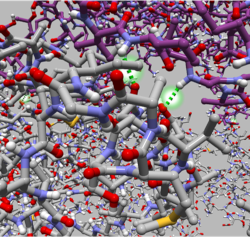 | 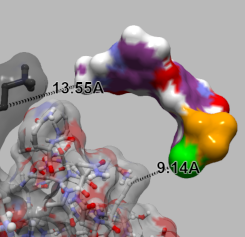 |
| GroEL and GroES. GroEL is rendered with ball-and-stick and semi-transparent molecular surface. GroES is rendered with the solid molecular surface. | The interatomic hydrogen bonds are depicted with green dashed lines. | Sorafenib and Braf. Dashed lines between atoms on each of the structures are shown to illustrate the distance between them. |
An OpenCL enabled GPU is required to run the software.
If you have any feedback or issues using DockIT please email: info@dockit.uk
For further information please visit the DockIT site by clicking here
If you have any difficulties downloading the DockIT tool, please contact us.
-
expand_more library_books References (7)
- Iakovou, G., Laycock, S.D. and Hayward, S. , Interactive Flexible-Receptor Molecular Docking in Virtual Reality Using DockIT, Journal of Chemical Information and Modeling, 2022
- Iakovou, G., Alhazzazi, M., Hayward, S. and Laycock, S.D. , DockIT: a tool for interactive molecular docking and molecular complex construction, Bioinformatics, 2020
- Matthews, N., Kitao, A., Laycock, S.D. and Hayward, S. , Haptic-assisted interactive molecular docking incorporating receptor flexibility, Journal of Chemical Information and Modeling, 2019
- Iakovou, G., Hayward, S. and Laycock, S.D. , Virtual Environment for Studying the Docking Interactions of Rigid Biomolecules with Haptics, Journal of Chemical Information and Modeling, 2017
- Iakovou, G., Laycock, S.D. and Hayward, S. , Determination of locked interfaces in biomolecular complexes using Haptimol_RD, Journal of Biophysics and Physicobiology, 2016
- Iakovou, G., Hayward, S. and Laycock, S.D. , Adaptive GPU-accelerated force calculation for interactive rigid molecular docking using haptics, Journal of Molecular Graphics and Modelling, 2015
- Iakovou, G., Hayward, S. and Laycock, S.D. , A real-time proximity querying algorithm for haptic-based molecular docking, Faraday Discussion 169, 2014
-
expand_more cloud_download Supporting documents (0)Additional files may be available once you've completed the transaction for this product. If you've already done so, please log into your account and visit My account / Downloads section to view them.